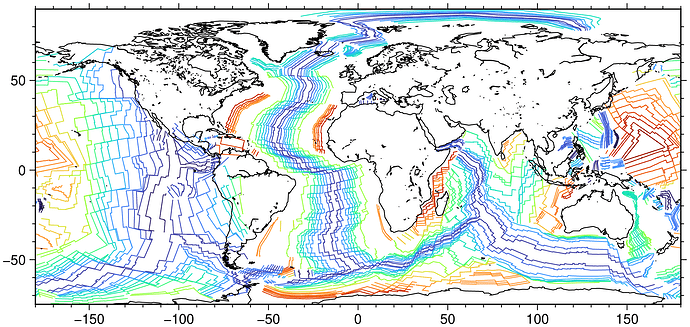
I am trying to plot isochrons that are exported from Gplates at several reconstructed times in my GMT maps, and I want to color code the isochrons based on their age. I know this info is in the Gplates export files, and I was hoping GMT would automatically figure it out when I supply a cpt using -C in gmt plot, ie something like:
gmt plot $isochrons -W0.005i,dashed -C$isochron_age_color.cpt -V
but this does not seem to work, they are all black. Looking at the Gplates exports, I see that the age is in the header row of each isochron in the isochron export file, but there is no Z column in the rows of lat lons for the isochron trace. Is there a way to indicate that the Z column should be taken from the header row in a certain position? For example, here is an isochron from one of the exports:
@D701|40|Seton_etal_2020_Isochrons.gpmlz|Zahirovic_etal_2022_OptimisedMantleRef_and_NNRMantleRef.rot|833|gpml:Isochron|IC|55.9|-999|“TASMAN SEA, LORD HOWE RISE-AUSTRALIA ANOMALY 25y ISOCHRON CG <identity”|GPlates-930671a3-e10c-4836-a3ab-72c6bebf7a3c|801|99|1|1423|25|5|37
151.911907382949 -44.5998189313693
152.718229716401 -45.3964628557164
152.532681479465 -45.6466035012743
153.053169106557 -46.3532948491846
152.910040550203 -46.6248766351768
153.138201981277 -46.8378356026436
154.263569422899 -47.6760773660758
The age of this particular example (55.9) is bolded in the header row but its a bit hard to see.
Many thanks for the help.
- Peter