In multiple datasets I am having challenges with corrupt PyGMT surface outputs related to the max_radius and registration options. In both cases the output grid has a diagonal line from lower left to upper right, and shifted and skewed grids that are discontinuous between upper left and lower right halves.
I almost always get the problem when using any setting for max_radius. If I leave out max_radius the problem still persists in some cases but can usually be resolved by switching from registration=g (gridline) to registration=p (pixel) or vice-versa.
In all cases I have applied blockmean to the input data and avoided prime grid cell numbers, and the problem occurs for tiny grids (~4000 data points) and large grids (3.3M data points). I have found that every now and then I get lucky and find a subset of the data where all settings work. The output of blockmean is not corrupted: it is definitely surface.
For grids that have problems, all max_radius settings fail (small or large, cell-based or distance based) and only one of gridline vs pixel will work.
4 examples are shown below for the same input data in standard UTM coordinates on NW-oriented lines:
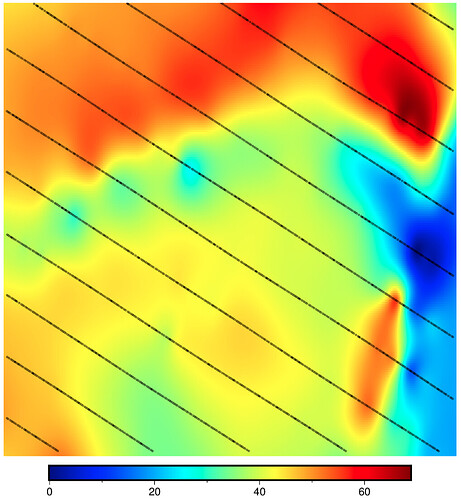
CORRECT RESULT: max_radius not used (M=None) and pixel registration (registration="p")
n_columns: 304 n_rows: 200
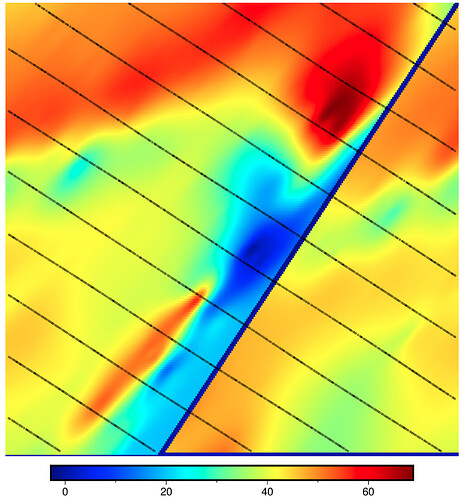
CORRUPT RESULT: max_radius not used (M=None) and gridline registration (registration="g")
n_columns: 305 n_rows: 201
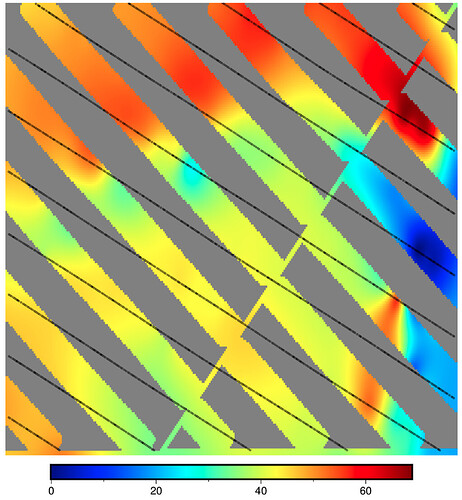
CORRUPT RESULT: max_radius = 5 cells (M="5c") and pixel registration (registration="p")
n_columns: 304 n_rows: 200
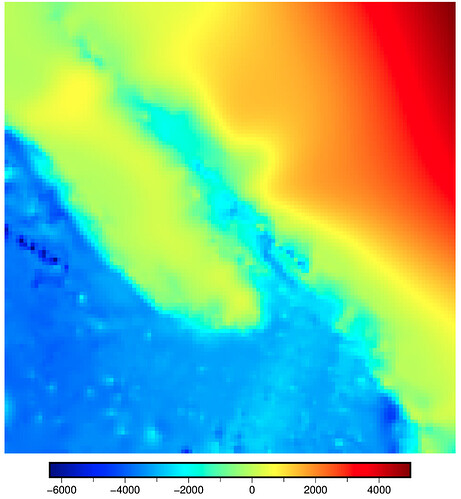
CORRUPT RESULT: max_radius = 5 cells (M="5c") and gridline registration (registration="g")
n_columns: 305 n_rows: 201
Any ideas how to avoid these problems and enable max_radius to be used? It looks like the coordinates have been transformed somehow, but then incorrectly transformed back?