Hi,
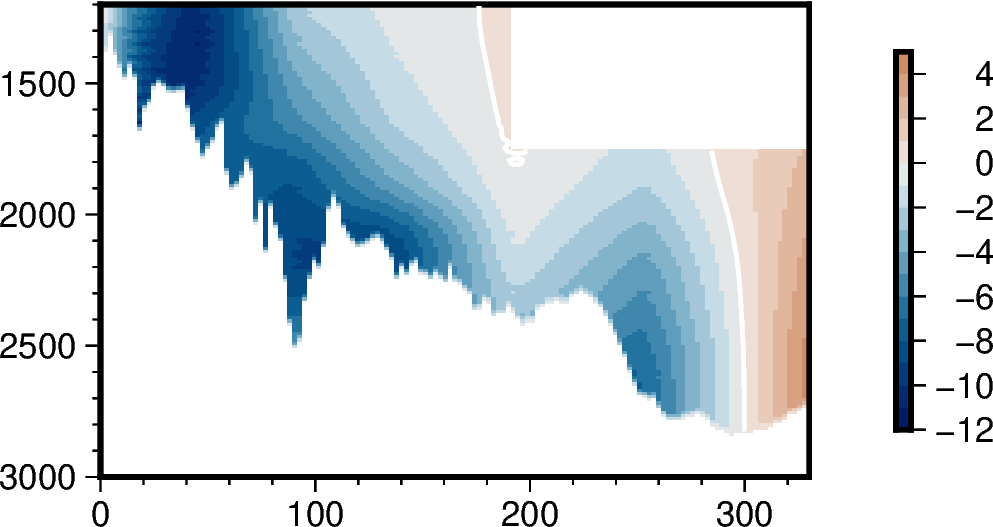
I’m trying to reproduce a cartesian matplotlib plot of a velocity section in pygmt, as I want to combine several of these with a pygmt map. This was produced using contourf(), but I’m struggled to get the same result with pygmt.
iceland_basin_velocity_section.pdf (47.8 KB)
I’ve tried grdimage(), which gives a gradient rather than the same color between contours. Can I use grdcontour() and a CPT? The data are diverging, and are asymmetric around zero. Do I need to make a diverging, asymmetric CPT? The GMT API shows a ‘-N’ option, which sounds like it might do what I want, but I’m not sure how to use this with the pygmt function, which doesn’t list an equivalent option.
If it’s not possible, please do let me know and I’ll use Inkscape to combine plots instead, as I’m new to PyGMT, it’s a very steep learning curve, and I’m very pressed for time at the moment!
many thanks for any help in advance,
Emma





 !
!