Hello everyone,
I am new to GMT and i cannot for the life of me find a solution to my problem. I writing a small code in julia where i read a netcdf dataset spanning over a year. When i extract the longitude and latitude data, i am able with the coast function to generate a map over the desired portion of earth (in this case Europe).
Then i add the grdimage! function in which i give him a grid thanks to the inbuilt GMT.mat2grid function. However, everytime i try, i only get the map of Europe, with no data on it.
Here is my code :
Summary
using NCDatasets
using GMT
println("")
println(“beginning”)
println("")
i=13 #this is to give me data at 12am
data=Dataset(“data_2009.nc”) #import my data
ssr=data[“ssr”]
lat=data[“latitude”]
lon=data[“longitude”]
time=data[“time”]
ssr=ssr.var[:,:].*ssr.attrib[“scale_factor”].+ssr.attrib[“add_offset”] #unfortunately NCDatasets doesn’t do
#any calculation and i need to do it
#myself
ssr=ssr[:,:,i] #get data from time i
lat=lat.var[:] #transform the data of lat into an array
lon=lon.var[:] #same with lon
minssr=minimum(ssr) #find minimum of ssr, not used here but will be later on in my code
maxssr=maximum(ssr) #same with maximum
minlat=minimum(lat) #finding minimum of lat in order to feed into GMT.mat2grid
maxlat=maximum(lat) #same
minlon=minimum(lon) #same
maxlon=maximum(lon) #same
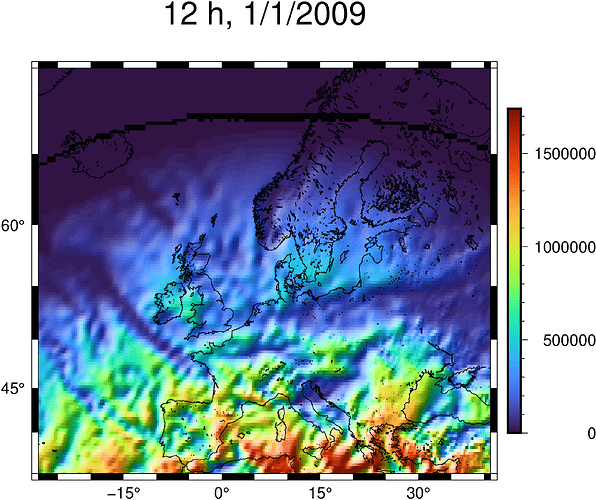
grid=GMT.mat2grid(ssr,x=collect(minlat:0.25:maxlat),y=collect(minlon:0.25:maxlon)) #making the grid
mycmap = makecpt(color=“blue@100,white@50,red”,continuous=true) #no idea if this fella works
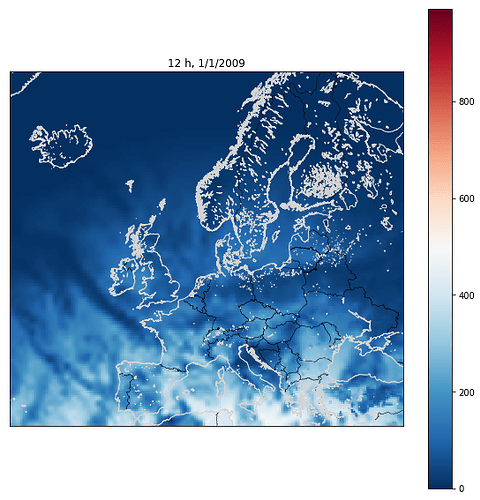
coast(region=(minlon,maxlon,minlat,maxlat), #this function works, return a map over Europe
proj=:Mercator,
res=:full,
noframe=:true,
axis=:none,
frame=:false,
land=:gray,
borders=1,
scale=1,
title=time[i])
grdimage!(grid,cmap=mycmap,dpi=200,t=50,show=true) #doesn’t work at all, no heatmap generated over
#coast
And when i run the code, the grid is done correctly as it shows me an array of 277x141 :
x_min: 35.0 x_max :70.0 x_inc :0.25 n_columns :141
y_min: -28.0 y_max :41.0 y_inc :0.25 n_rows :277
z_min: 0.0 z_max :1.7404528786412952e6
However i do get this error everytime,
pscoast [ERROR]: Bad interval in -B option (x-component, a-info): lse gave interval = 0
pscoast [ERROR]: Bad interval in -B option (y-component, a-info): lse gave interval = 0
[:noframe, :axis]
I tried different options with xaxis and yaxis, but i need to admit, i don’t know what i am doing at this point.
Regards