Hi All -
I have been plotting .grd files and for some reason the highest latitude position in my file are either not being read or are just not plotting. I know that the data in the file are valid and are within the cpt I made for these plots. Here is a small sample of the .grd file.
-114.40000 62.20000 4.028
-114.20000 62.20000 4.028
-114.00000 62.20000 4.028
-113.80000 62.20000 4.028
-113.60000 62.20000 4.028
-113.40000 62.20000 4.028
-113.20000 62.20000 4.028
-113.00000 62.20000 4.028
-112.80000 62.20000 3.842
-114.40000 62.40000 4.028
-114.20000 62.40000 4.028
-114.00000 62.40000 3.842
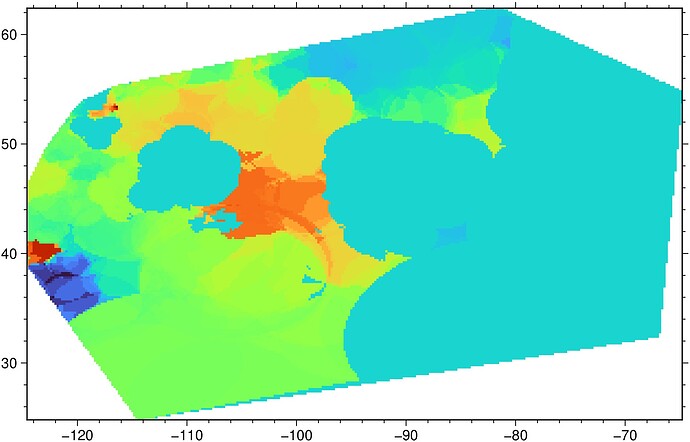
This lon/lat is up around Yellowknife in central Canada.
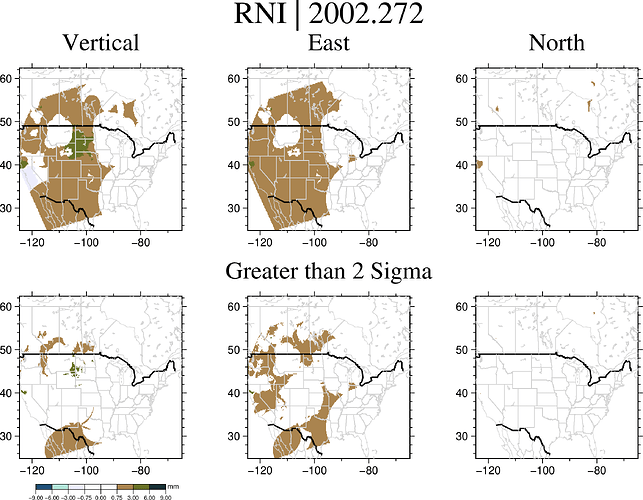
As an example, here is one of the plots. You can see that there is a line that cuts below where that the above lon/lat is specified – and that is true for every plot I make even though there is data that should be plotted around the Yellowknife area. I can’t seem to figure out why this is happening.
To the best of my knowledge, I’m not clipping any regions, and I’ve specified that my region is 2-degrees higher at 64-degree latitude to ensure that my plot would include the entirety of my data.
Here is the section of the .grd file for the plot shown above, which should, again, be shown in the plot, but is absent:
-114.40000 62.20000 1.088
-114.20000 62.20000 1.088
-114.00000 62.20000 1.088
-113.80000 62.20000 1.088
-113.60000 62.20000 1.088
-113.40000 62.20000 1.088
-113.20000 62.20000 1.088
-113.00000 62.20000 1.088
-112.80000 62.20000 0
-114.40000 62.40000 1.088
-114.20000 62.40000 1.088
-114.00000 62.40000 1.088
And here is the code I have written to create the above plot:
GMT.subplot(grid = "2x3",
title = "RNI | $yr",
clearance = (top = 0.5),
savefig = imgpath * "$count_up" * ".png");
# rad
GMT.grdimage(sigrad, (region= [-130 -70 24 64], proj=:Winkel),
coast = (borders = ((type = 1, pen = ("thick", "black")), (type = 2, pen = ("thinner", "lightgrey"))),
area = 500,
shore = (:thinnest, :lightgrey)),
cmap = bukavu,
panel = (1,1),
title = "Vertical"
);
GMT.grdimage(sigsigrad, (region= [-130 -70 24 64], proj=:Winkel),
coast = (borders = ((type = 1, pen = ("thick", "black")), (type = 2, pen = ("thinner", "lightgrey"))),
area = 500,
shore = (:thinnest, :lightgrey)),
cmap = bukavu,
panel = (2,1)
);
# lon
GMT.grdimage(siglon, (region= [-130 -70 24 64], proj=:Winkel),
coast = (borders = ((type = 1, pen = ("thick", "black")), (type = 2, pen = ("thinner", "lightgrey"))),
area = 500,
shore = (:thinnest, :lightgrey)),
cmap = bukavu,
panel = (1,2),
title = "East"
);
GMT.grdimage(sigsiglon, (region= [-130 -70 24 64], proj=:Winkel),
coast = (borders = ((type = 1, pen = ("thick", "black")), (type = 2, pen = ("thinner", "lightgrey"))),
area = 500,
shore = (:thinnest, :lightgrey)),
cmap = bukavu,
panel = (2,2),
title = "Greater than 2 Sigma"
);
# lat
GMT.grdimage(siglat, (region= [-130 -70 24 64], proj=:Winkel),
coast = (borders = ((type = 1, pen = ("thick", "black")), (type = 2, pen = ("thinner", "lightgrey"))),
area = 500,
shore = (:thinnest, :lightgrey)),
cmap = bukavu,
panel = (1,3),
title = "North"
);
GMT.grdimage(sigsiglat, (region= [-130 -70 24 64], proj=:Winkel),
coast = (borders = ((type = 1, pen = ("thick", "black")), (type = 2, pen = ("thinner", "lightgrey"))),
area = 500,
shore = (:thinnest, :lightgrey)),
cmap = bukavu,
panel = (2,3)
);
colorbar!(position=(outside=true, anchor=:BC), equal=true, cmap=bukavu, ylabel = "mm");
Here is a link for the data via proton drive, if you would like to check out the complete data for this epoch.
If that link doesn’t work for any reason, feel free to PM me and I can send you the data via email if you’re so inclined.
Thank you for any help or insight ![]()
- Rob
EDIT: The fact that the columns I copy & pasted are showing up red in this seems weird. Could the reason behind the columns being shown in red here be a clue to as to why my data is not plotting? I’m not sure why those would be a problem though. The rest of the columns are formatted the same way, and the lon/lat are within my map region too. ![]()