Hello PyGMT Forum,
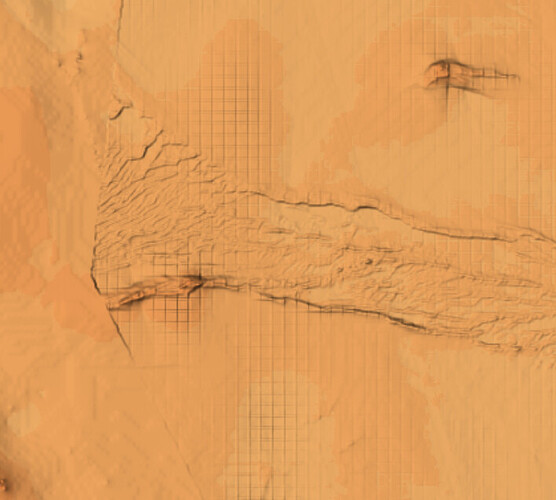
I have provided my code script and a picture of the grid issue I’m getting from the output of the nearest neighbor method. Basically I’m combining a number of XYZ datasets in a duckdb database, ingesting it as a GDF, then using the block median and then nearest neighbor methods to create the grid. I am getting issues when I merge all datasets as pictured below. I thought the block median method would create an evenly distributed grid but when i plot the output on the map it is not evenly distributed in the areas where I’m getting the grid issues. When I perform this process of the XYZ datasets individually it is not occurring, only on the merged datasets. My data is in EPSG4326 crs.
I cannot figure out what I’m doing wrong or am I interpretting the way the block median method works incorrectly?
Grid issue when datasets merged prior to gridding
How grid looks when gridded individually and overlaid
Example of blockmedian output as points on map
Example of script below
import duckdb
import os
import geopandas as gpd
import warnings
import pandas as pd
import pygmt
con = duckdb.connect(r"D:\QLD\DB\QLD.db")
existing_table_name = "qld_test_merge_v2"
out_file_name = "test_merge"
geom_col = 'geom'
min_x = 150 # west
max_x = 151 # east
min_y = -21 # south
max_y = -20 # north
new_table = "test_merge_new"
# Install and load the 'spatial' extension
con.execute("INSTALL 'spatial';")
con.execute("LOAD 'spatial';")
# Create a new table with the filtered data
#con.execute(f"CREATE OR REPLACE TABLE {new_table} AS SELECT * FROM {existing_table_name} WHERE X >= {min_x} AND X <= {max_x} AND Y >= {min_y} AND Y <= {max_y}")
# Read database as GeoDataFrame
try:
gdf = gpd.GeoDataFrame.from_postgis("SELECT * FROM "+existing_table_name, con, geom_col=geom_col)
except:
gdf = pd.read_sql("SELECT * FROM "+existing_table_name, con)
gdf = gdf.astype(float)
# Convert dataframe to geodataframe
gdf_convert = gpd.GeoDataFrame(gdf, geometry=gpd.points_from_xy(gdf.X, gdf.Y), crs="EPSG:4326")
gdf_convert = gdf_convert.rename(columns={"geometry": "geom"})
gdf = gdf_convert
print(gdf)
if 'X' not in gdf.columns:
gdf['X'] = gdf['geom'].x
if 'Y' not in gdf.columns:
gdf['Y'] = gdf['geom'].y
#gdf = gdf[(gdf['X'] >= bounding_box[0])&(gdf['X'] <= bounding_box[1])&(gdf['Y'] >= bounding_box[2])&(gdf['Y'] <= bounding_box[3])]
gdf = gdf.reset_index()
gdf = gdf[['geom','X','Y','Z']]
print(gdf)
# Get the min and max X and Y
xmin = gdf['X'].min()
xmax = gdf['X'].max()
ymin = gdf['Y'].min()
ymax = gdf['Y'].max()
print(xmin, xmax, ymin, ymax)
grid = pygmt.blockmedian(
x=gdf.X,
y=gdf.Y,
z=gdf.Z,
spacing="30e",
region=[xmin, xmax, ymin, ymax],
#outfile=grid_file
)
col_names = {0: 'X', 1: 'Y', 2: 'Z'}
grid.rename(columns=col_names, inplace=True)
print("Block median operation for completed successfully.")
print(grid)
outpath1 = out_file_name+"_nn.nc"
outpath2 = out_file_name+"_filter.nc"
#xmin2 = grid['X'].min()
#xmax2 = grid['X'].max()
#ymin2 = grid['Y'].min()
#ymax2 = grid['Y'].max()
output = pygmt.nearneighbor(
data=grid,
spacing="30e",
region=[xmin, xmax, ymin, ymax],
search_radius="90e",
nodata="0",
outgrid=outpath1
)
print("Grid completed successfully")
'''
pygmt.grdfilter(
grid=outpath1,
outgrid=outpath2,
filter="c0.0015",
distance="0.003",
#region=[xmin, xmax, ymin, ymax],
spacing="0.0003",
x=16,
verbose=True
)
'''
#con.sql("DROP TABLE "+new_table)
#print("table dropped"+new_table)
print("Filter completed successfully")