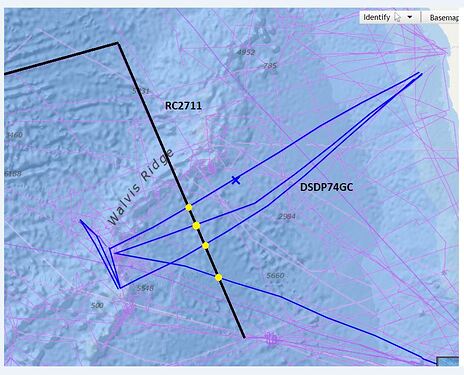
Hey there!
I am new to pygmt. I am trying to calculate the intersections between two ship tracks using x2sys_cross.
The tracks have 4 points of intersection but x2sys_cross outputs only one intersection.
I wrote the code to calculate the intersection following the code posted on another question in the forum.
# running x2sys_init
pygmt.x2sys_init(
tag="mag",
fmtfile="xyzt",
suffix="xyzt",
discontinuity="d", # geographical coordinates discontinuity at dataline
spacing="10m",
units=["dk", "sn"], # distance in kilometers, speed in knots
gap="d1k", # distance gap up to 1 km allowed
force=True, # replace old files
verbose="w"
)
# reading ship tracks as dataframes
track1: pd.DataFrame = pd.read_csv('dsdp74gc.xyzt',sep='\t',skiprows=1) # 1st file with x, y, z, t columns
track2: pd.DataFrame = pd.read_csv('rc2711.xyzt',sep='\t',skiprows=1) # 2nd file with x, y, z, t columns
# running x2sys_cross
df_crossover: pd.DataFrame = pygmt.x2sys_cross(
tracks=[track1, track2],
tag="mag",
interpolation="l", # linear interpolation
coe="e", # external crossovers
speed=["l1", "u15", "h1"],
#trackvalues=False,
trackvalues=True # Get track 1 height (h_1) and track 2 height (h_2)
)
# Results of Cross over
print (df_crossover)
lon lat t_1 t_2 dist_1 \
0 6.041174 -28.864184 1980-06-25 12:18:54 1986-12-07 14:39:15 1472.200472
dist_2 head_1 head_2 vel_1 vel_2 z_1 z_2
0 137.521438 20.389965 293.781108 8.29052 10.143803 79.679097 96.833434
Maybe I’m missing something. Can anyone please help?
This zip folder has the script, input data and the format definition file.
data.zip (130.4 KB)
Appreciate any leads!