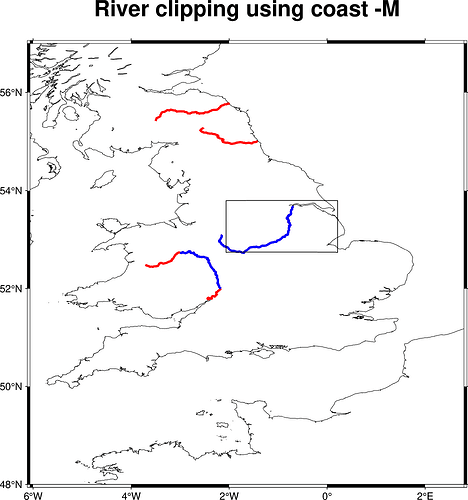
One apparent possibility is to use clip, the other to dump data using coast -M
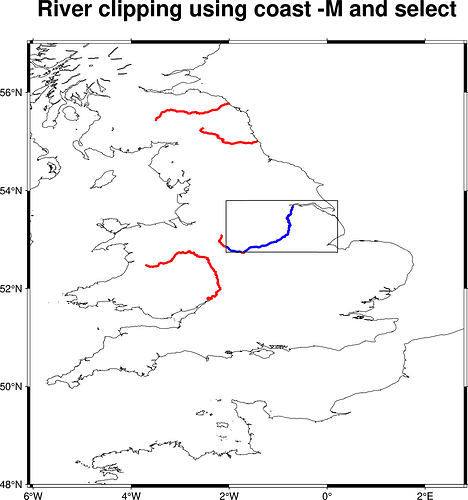
Neither appears to be wrapped by pygmt as far as I can understand. I tried using both using GMT C API calls mechanism provided by pygmt, just out of curiosity. clip does the job precisely but the result looks ugly as clip clips the graphic representation which does not look good if rivers were plotted using a thick pen. coast however dumped an extra river segment quite far outside the specified -R... boundaries which does not look too good either. See some spagetti code and the plots below. Blue line shows rivers clipped out by the corresponding method. Red line shows all level 3 rivers -I3/2p,blue,solid as specified by the topic started. Why does coast behave so? Thanks!
using psclip:
import pygmt
XMIN=-2.062697; XMAX=0.214867; YMIN=52.737217; YMAX=53.791475
clipping_rectangle=f'{XMIN} {YMIN}\n{XMIN} {YMAX}\n{XMAX} {YMAX}\n{XMAX} {YMIN}\n{XMIN} {YMIN}\n'
clipping_rectangle_file = 'clipping_rectangle.txt'
with open(clipping_rectangle_file,"w") as f:
f.write(clipping_rectangle)
fig = pygmt.Figure()
region = '-6.061164/2.790309/48/57'
title = 'River clipping using psclip'
fig.coast(
region=region,
shorelines=True,
rivers='3/2p,red,solid',
frame=[f'WSen+t{title}',"xa","ya"]
)
# equivalent to
# gmt clip -R-2.062697/0.214867/52.737217/53.791475 clipping_rectangle.txt
with pygmt.clib.Session() as s:
# Initialize clipping
args = [f'{clipping_rectangle_file}', f'-R{region}']
s.call_module('clip', args=' '.join(args))
fig.coast(
#region='-2.062697/0.214867/52.737217/53.791475',
rivers='3/2p,blue,solid',
)
with pygmt.clib.Session() as s:
# Finalize clipping
s.call_module('clip', args='-C1')
fig.plot(
region=region,
data=clipping_rectangle_file,
pen='0.5p,black,solid'
)
fig.savefig("clipping_pygmt_1.png", show=True)
using coast -M:
import pygmt
XMIN=-2.062697; XMAX=0.214867; YMIN=52.737217; YMAX=53.791475
clipping_rectangle=f"{XMIN} {YMIN}\n{XMIN} {YMAX}\n{XMAX} {YMAX}\n{XMAX} {YMIN}\n{XMIN} {YMIN}\n"
clipping_rectangle_file = 'clipping_rectangle.txt'
with open(clipping_rectangle_file,"w") as f:
f.write(clipping_rectangle)
fig = pygmt.Figure()
region='-6.061164/2.790309/48/57'
title = 'River clipping using coast -M'
fig.coast(
region=region,
shorelines=True,
rivers='3/2p,red,solid',
frame=[f'WSen+t{title}',"xa","ya"]
)
region1 = '-2.062697/0.214867/52.737217/53.791475'
clipped_river_file = 'clipped_river_file.txt'
# equivalent to
# gmt coast -R-2.062697/0.214867/52.737217/53.791475 -M -I3 > clipped_river_file.txt
with pygmt.clib.Session() as s:
args = [f'-R{region1}', '-M', '-I3', f'> {clipped_river_file}']
s.call_module('coast', args=' '.join(args))
fig.plot(
region=region,
data=clipped_river_file,
pen='2p,blue,solid'
)
fig.plot(
region=region,
data=clipping_rectangle_file,
pen='0.5p,black,solid'
)
fig.savefig("clipping_pygmt_2.png", show=True)