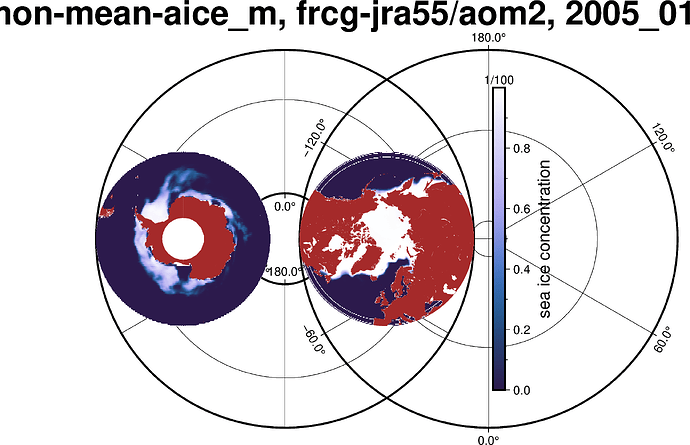
Hello,
I am currently using Python (and associated libraries: numpy, xarray, matplotlib, cartopy, proplot) to plot the following figure.
The issue I am facing is that it is taking roughly 90 seconds to do this. As an alternative I would like to use pygmt as it may be faster. I have used GMT and PyGMT before for figure creation and have appreciated its quality and speed. However, when I attempt to plot this grid using grdimage I run into an irregular grid problem, then when I attempt to use grd2xyz it keys on the dimensions ‘nj’ and ‘ni’ but not the coordinates. Then when I swap the ‘nj’ and ‘ni’ dimensions out to their corresponding geographic lon/lat dimensions and run through grd2xyz I get a kernel panic / crash.
Curious to hear any suggestions for a work around.
Here is the result of grdinfo:
<xarray.DataArray ‘aice_m’ (nj: 1080, ni: 1440)>
array([[nan, nan, nan, …, nan, nan, nan],
[nan, nan, nan, …, nan, nan, nan],
[nan, nan, nan, …, nan, nan, nan],
…,
[nan, nan, nan, …, nan, nan, nan],
[nan, nan, nan, …, nan, nan, nan],
[nan, nan, nan, …, nan, nan, nan]], dtype=float32)
Coordinates:
time object 2005-02-01 00:00:00
TLON (nj, ni) float32 79.88 80.12 80.38 80.62 … 80.0 80.0 80.0 80.0
TLAT (nj, ni) float32 -81.08 -81.08 -81.08 -81.08 … 65.34 65.24 65.13
ULON (nj, ni) float32 -280.0 -279.8 -279.5 -279.2 … 80.0 80.0 80.0
ULAT (nj, ni) float32 -81.02 -81.02 -81.02 -81.02 … 65.29 65.18 65.08
Dimensions without coordinates: nj, ni
Attributes:
units: 1
long_name: ice area (aggregate)
cell_measures: area: tarea
cell_methods: time: mean
time_rep: averaged
grdinfo [WARNING]: “nj”, NetCDF: Variable not found
If something bad happens later, try importing via GDAL.
grdinfo [WARNING]: “ni”, NetCDF: Variable not found
If something bad happens later, try importing via GDAL.
grdinfo [WARNING]: “d2”, NetCDF: Variable not found
If something bad happens later, try importing via GDAL.
: Title:
: Command:
: Remark:
: Pixel node registration used [Cartesian grid]
: Grid file format: nf = GMT netCDF format (32-bit float), CF-1.7
: x_min: -0.5 x_max: 1439.5 x_inc: 1 name: x n_columns: 1440
: y_min: -0.5 y_max: 1079.5 y_inc: 1 name: y n_rows: 1080
: v_min: 0 v_max: 0.999973714352 name: z
: scale_factor: 1 add_offset: 0
: format: classic
: Default CPT:
da = xr.load_dataset('./iceh.2005-01.nc',chunks=None).aice_m.isel(time=0)
da['longitude'] = xr.DataArray(data=da.TLON.data[0,:],dims='ni').set_index()
da['latitude'] = xr.DataArray(data=da.TLAT.data[:,0],dims='nj').set_index()
da = da.drop(['ULAT','ULON','TLON','TLAT'])
#da = da.swap_dims({'nj':'latitude','ni':'longitude'})
print(da)
print(pygmt.grdinfo(grid=da))
fig = pygmt.Figure()
fig.coast(region = [0, 360, -90, -50], projection = "S0.0/-90.0/50/5i", shorelines = True)
fig.basemap(frame = ["a", "WSNE"])
#xyz = pygmt.grd2xyz(grid=da)
fig.grdimage(grid=da,cmap= "haxby",frame=True)
fig.show()