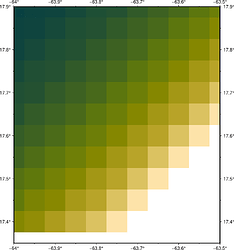
Is is possible to replicate gmt grdview -T with pygmt? I want to see the pixel values without interpolation (pixel outlines optional).
Scripts and sample data below. PNG versions included inline.
Question: gmt grdview -T shows each pixel without interpolation while pygmt.grdview(...) interpolates pixel values (and seems to draw contours). Can interpolation be turned off with pygmt.grdview()? Is there a different way to achieve this?
With regular GMT:
#!/bin/bash
gmt begin test_shell
gmt set COLOR_BACKGROUND dimgray COLOR_FOREGROUND gray
gmt set MAP_FRAME_TYPE plain FORMAT_GEO_MAP D
RANGE=$(gmt info demo-data.txt -I0.05)
PROJ="-JM6i"
gmt xyz2grd -Gdemo-data.grd demo-data.txt -I0.05 $RANGE $PROJ
gmt makecpt -Cbamako -T1e-8/1/1+l -Ic -Z
gmt grdview -T demo-data.grd -C -BNSEW -Ba.1f.05
gmt end show
With pygmt:
import pygmt
txt = "demo-data.txt"
range = pygmt.info(txt, spacing=0.05)
proj = "M6i"
grd = pygmt.xyz2grd(txt, spacing=0.05, region=range)
fig = pygmt.Figure()
with pygmt.config(COLOR_BACKGROUND="dimgray", COLOR_FOREGROUND="gray", COLOR_NAN="white"):
pygmt.makecpt(cmap="bamako", series="1e-8/1/1+l", reverse=True, continuous=True, overrule_bg=True)
fig.grdview(
grd,
# drapegrid=grd,
surftype="s",
frame=["a.1f.05", "NSEW"]
)
fig.savefig("test_pygmt.pdf", crop=True, dpi=300, show=True)
demo-data.txt (2.9 KB)
Any other suggestions to improve my pygmt script are welcome!
Thank you,
Jason
GMT v. 6.5.0, pygmt v. 0.12.0, macOS 13.6.7, vscode
a call to GMT C level api works for me:
...
#fig.grdview(
# grd,
# # drapegrid=grd,
# surftype="s",
# frame=["a.1f.05", "NSEW"]
#)
with pygmt.clib.Session() as s:
with s.virtualfile_in(check_kind="raster", data=grd) as vingrd:
args = [f'{vingrd}', '-T', '-BNSEW', '-Ba.1f.05', '-C']
s.call_module('grdview', args=' '.join(args))
...
2 Likes
Wow ! that’s a very interesting way to explore. Is there any tutorial on how to use C level inside pygmt ?
1 Like
No, I don’t think there is a tutorial, I haven’t seen any. I read pygmt C api documentation API Reference — PyGMT and pygmt.clib.Session.call_module — PyGMT and browsed through pygmt sources for modules on more practical examples of its use.
In principle calling gmt modules is simple, like call_module above runs a gmt command with name specified as string with the parameters specified as the args string. Calling GMT commands one by one for working with datafiles like in plain GMT requires nothing extra, just passing options as strings same way as when using CLI gmt.
it becomes more complex when one needs to parse output from commands like gmt info and use data produced inside the python script. For that pygmt people developed a virtual file mechanism see e.g. pygmt.clib.Session.virtualfile_in — PyGMT and other types of virtualfile calls.
2 Likes
Wow! I didn’t know about clib. Is it possible to use the clib session methods and pygmt.Figure methods (e.g. fig.coast()) to build a single figure? Or would you need to use one or the other? All session.call_module(...) within a pygmt.clib.Session() context, or all pygmt Figure methods.
I’ll have to read up on clib and do some experimenting - it could be a great way to finally switch my workflows from shell scripts to Python.
1 Like
grdview’s -T option is not implemented in PyGMT yet, but you can use Figure.grdview(grid, surftype="s", T=True) as a temporary workaround.
1 Like
I have to omit surftype, otherwise it throws an error: “Options -Q and -T are mutually exclusive”. But T=True works.
Thanks all!
1 Like